Interaction lists
In the BioRECIPE format, interactions can be represented using the event-based interaction list format where each individual interaction (biological event) is assigned one row. The column headers match interaction attribute names.
An example biological interaction, represented as a directed signed edge between two nodes, including node, edge, context, and provenance attributes is illustrated in the figure below (subscripts: s - source node, t - target node, e - edge).

An illustration of the interaction list in the BioRECIPE format, with all the attributes (current version supports one source node and one target node in each interaction). See an example file here.
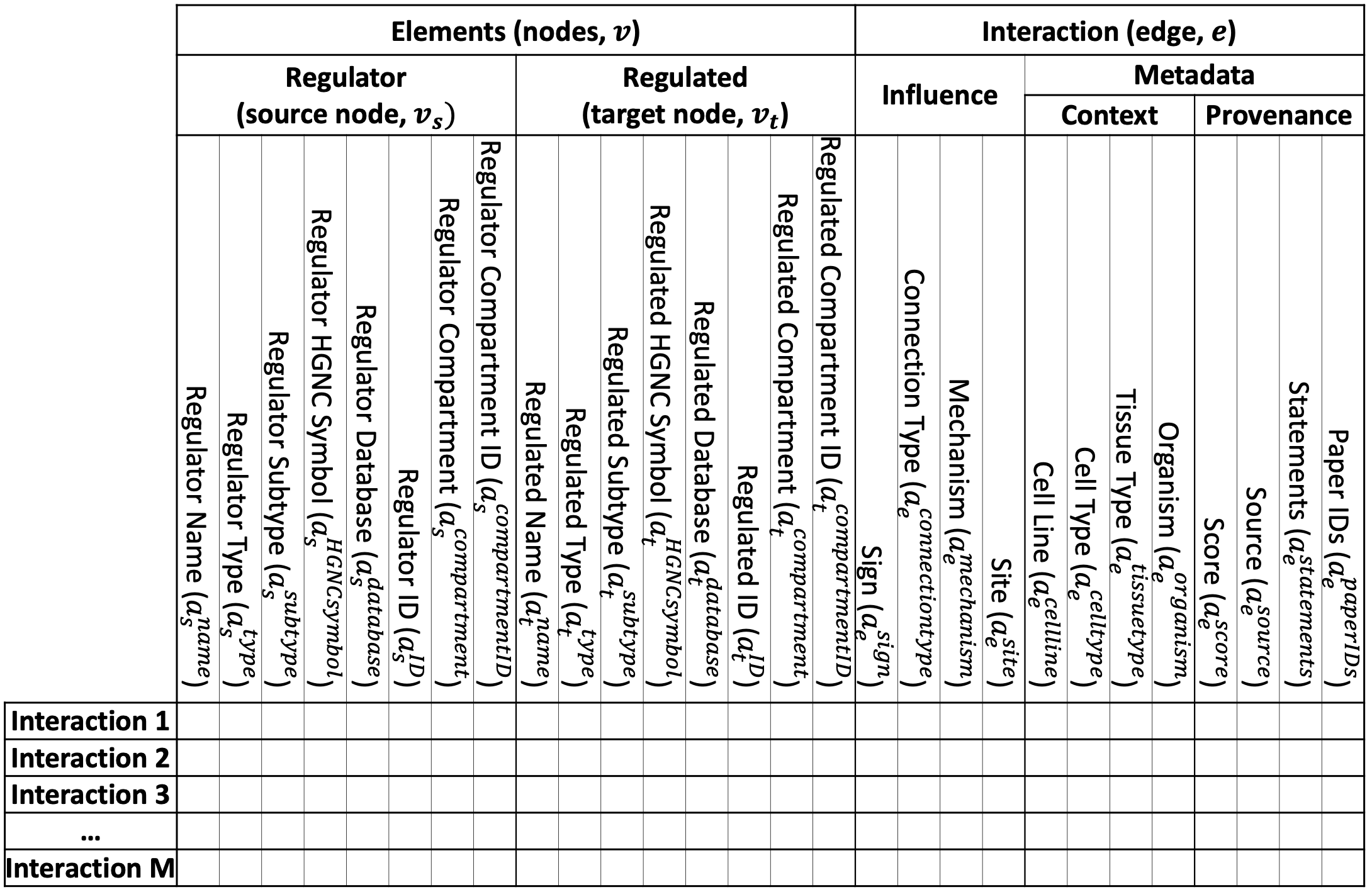
As shown in the table above, the BioRECIPE representation of an interaction allows for many interaction attributes to be included. Each attribute belongs to one of the following groups:
basic element attributes
basic interaction attributes
context attributes
provenance attributes
The following tables provide details for each attribute, including attribute name used in the BioRECIPE spreadsheet, a symbol used in detailed definitions, a brief description of the attribute, allowed values, and a few examples. Additional interaction examples can be found here.
Basic element attributes
Attribute |
Symbol |
Description |
Values |
Examples |
|---|---|---|---|---|
Name |
\(a^{\mathrm{name}}\) |
a common name of a biological component used by experts |
<element name> |
ERK1, RAS, p53 |
Type |
\(a^{\mathrm{type}}\) |
biological component type |
|
listed under Values |
Subtype |
\(a^{\mathrm{subtype}}\) |
element subtype provides additional details for curation |
<subtype name> |
receptor, kinase, transcription factor |
HGNC Symbol |
\(a^{\mathrm{HGNCsymbol}}\) |
the corresponding gene symbol from the HGNC database |
<HGNC unique gene symbol> |
BCL2L1, APAF1 |
Database |
\(a^{\mathrm{database}}\) |
a database where the element ID is found |
|
listed under Values |
ID |
\(a^{\mathrm{ID}}\) |
unique element ID from an open access database |
<unique identifier> {, <unique identifier>} |
Q07817, O14727 |
Compartment |
\(a^{\mathrm{compartment}}\) |
cellular compartment name |
|
listed under Values |
Compartment ID |
\(a^{\mathrm{compartmentID}}\) |
cellular compartment unique identifier from the GO database |
|
identifiers are listed under Values in the same order as compartment names |
Basic interaction attributes
Attribute |
Symbol |
Description |
Values |
Examples |
|---|---|---|---|---|
Direction |
\(a^{\mathrm{direction}}\) |
this is an implicit attribute |
determined as a direction from source to target node |
|
Sign |
\(a^{\mathrm{sign}}\) |
interaction sign (also referred to as “polarity”) indicates positive or negative influence |
|
listed under Values |
Connection Type |
\(a^{\mathrm{connectiontype}}\) |
indicates whether the edge between the source and target nodes represents direct physical interaction between elements, or it is expected or known that there is a path of several connected interactions between the source node and target node |
|
listed under Values |
Mechanism |
\(a^{\mathrm{mechanism}}\) |
indicates the exact physical interaction, i.e., biological mechanism; value usually included when the Connection Type is direct |
|
listed under Values |
Site |
\(a^{\mathrm{site}}\) |
molecular site where the interaction occurs |
<molecular site name> |
T308, T450, S473 |
Context attributes
Attribute |
Symbol |
Description |
Values |
Examples |
|---|---|---|---|---|
Cell Line |
\(a^{\mathrm{cellline}}\) |
cell line where the interaction is observed |
<cell line name> |
GS6-22 (glioblastoma multiforme (GBM) cell line) |
Cell Type |
\(a^{\mathrm{celltype}}\) |
cell type where the interaction is observed |
<cell type name> |
T cell, macrophage, pancreatic cancer cell, GBM cell |
Tissue Type |
\(a^{\mathrm{tissuetype}}\) |
tissue type where the interaction is observed |
<tissue type name> |
pancreas, colon, brain |
Organism |
\(a^{\mathrm{organism}}\) |
organism where the interaction is observed |
<organism name> |
human, mouse |
Provenance attributes
Attribute |
Symbol |
Description |
Values |
Examples |
|---|---|---|---|---|
Score |
\(a^{\mathrm{score}}\) |
confidence in interaction |
<number> |
a number in the interval [0,1] for sources like INDRA or STRING; present/absent for PCnet |
Source |
\(a^{\mathrm{source}}\) |
knowledge or data source where the interaction is found |
|
listed under Values |
Statements |
\(a^{\mathrm{statements}}\) |
statements (sentences) where the interaction is found |
<text>; {<text>;} |
“Bcl-XL interacts with Apaf-1 and inhibits Apaf-1-dependent caspase-9 activation” |
Paper IDs |
\(a^{\mathrm{paperIDs}}\) |
if literature, paper IDs where the interaction is found |
<PMCID | PMID> {,<PMCID | PMID>} |
PMID9539746 |
Other attribute definitions
Name |
Definition |
|---|---|
<element name> |
user-defined, typically a commonly used name for the biological species or component |
<subtype name> |
user-defined, used to specify element beyond just its type |
<HGNC unique gene symbol> |
a gene symbol from the HGNC database that corresponds to the element |
<molecular site name> |
commonly used molecular site symbol |
<cell line name> |
assigned name of a cell line |
<cell type name> |
commonly used cellular type name |
<tissue type name> |
commonly used tissue type name |
<organism name> |
commonly used organism name |
<PMCID | PMID> |
unique paper PMCID or PMID |
<number> |
just a number |
<text> |
any text |