Executable models
The BioRECIPE format supports representation of the static graph structure of models, as well as attributes necessary to study the dynamics. Models are represented in the BioRECIPE format using element-based approach.
A toy example of a model graph, including input and output nodes, directed edges, paths, feedback and feedforward loops, and cellular compartments is shown in the introduction.
Model formats
In the BioRECIPE format, each element in a model is assigned a row in the model table/spreadsheet. Different from event-based representation of interactions, in this element-based representation interactions in which the element participates as a regulated element (target node of interaction edge) can be combined into an element update rule. The BioRECIPE format supports several different model representation schemes, ranging from less detailed to more detailed, from static graph attributes to dynamic attributes and parameters necessary for analysis of dynamic behavior, as illustrated in the table below (x indicates attributes included in the representation scheme):
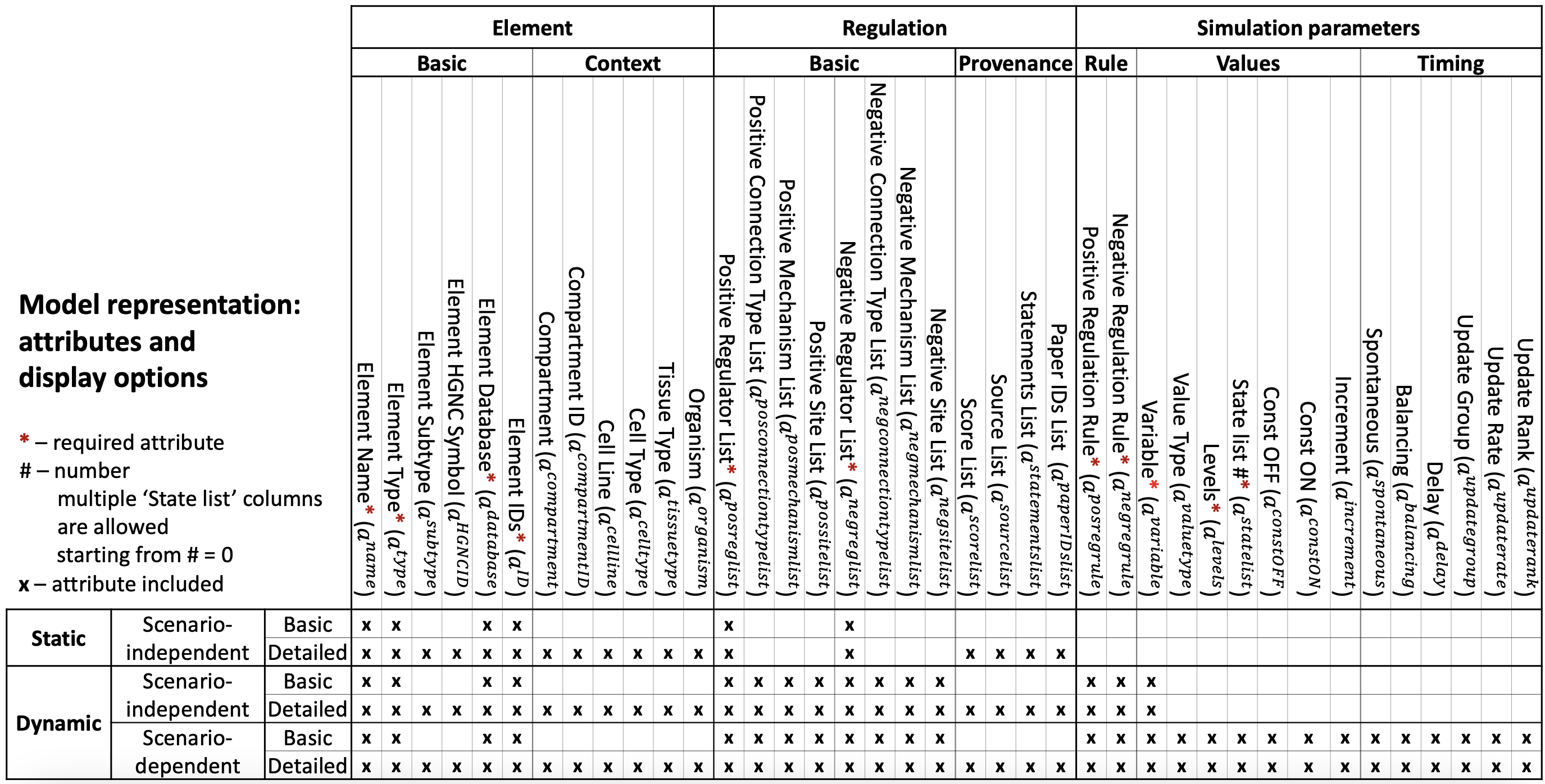
Model attributes
The following tables provide details for each attribute, including attribute name used in the BioRECIPE spreadsheet, a symbol used in detailed definitions, a brief description of the attribute, its allowed values, and a few examples for the attribute. Model examples can be found here.
The basic element (node) attributes, and context attributes are inherited from the interactions in which the element participates (as a regulated element), and they are included in the element representation, as shown in the figure above (columns under Element header).
Several other interaction attributes are ordered lists of values from all individual interactions where the element is a regulated element (target node on the interaction edge). These are referred to as Regulation attributes and include basic and provenance attributes combined from indvidual interactions (basic interaction attributes and provenance attributes).
Attribute |
Symbol |
Description |
Values |
Examples |
|---|---|---|---|---|
Positive regulator list |
\(a^{\mathrm{posreglist}}\) |
All positive regulators of the element from individual interactions, combined into a list |
<positive regulator 1>, <positive regulator 2>, …, <positive regulator \(k\)> |
examples |
Positive connection type list |
\(a^{\mathrm{posconnectiontypelist}}\) |
All connection types of element’s positive regulators from individual interactions, combined into a list, following the same order as in the positive regulator list |
<positive connection type 1>, <positive connection type 2>, …, <positive connection type \(k\)> |
examples |
Positive mechanism list |
\(a^{\mathrm{posmechanismlist}}\) |
All mechanisms of element’s positive regulators from individual interactions, combined into a list, following the same order as in the positive regulator list |
<positive mechanism 1>, <positive mechanism 2>, …, <positive mechanism \(k\)> |
examples |
Positive site list |
\(a^{\mathrm{possitelist}}\) |
All interaction sites of element’s positive regulators from individual interactions, combined into a list, following the same order as in the positive regulator list |
<positive site 1>, <positive site 2>, …, <positive site \(k\)> |
examples |
Negative regulator list |
\(a^{\mathrm{negreglist}}\) |
All negative regulators of the element from individual interactions, combined into a list |
<negative regulator 1>, <negative regulator 2>, …, <negative regulator \(l\)> |
examples |
Negative connection type list |
\(a^{\mathrm{negconnectiontypelist}}\) |
All connection types of element’s negative regulators from individual interactions, combined into a list, following the same order as in the negative regulator list |
<negative connection type 1>, <negative connection type 2>, …, <negative connection type \(l\)> |
examples |
Negative mechanism list |
\(a^{\mathrm{negmechanismlist}}\) |
All mechanisms of element’s negative regulators from individual interactions, combined into a list, following the same order as in the negative regulator list |
<negative mechanism 1>, <negative mechanism 2>, …, <negative mechanism \(l\)> |
examples |
Negative site list |
\(a^{\mathrm{negsitelist}}\) |
All interaction sites of element’s negative regulators from individual interactions, combined into a list, following the same order as in the negative regulator list |
<negative site 1>, <negative site 2>, …, <negative site \(l\)> |
examples |
Attribute |
Symbol |
Description |
Values |
Examples |
|---|---|---|---|---|
Score list |
\(a^{\mathrm{scorelist}}\) |
all scores of element’s positive and negative regulations from individual interactions, combined into a list, following the order of positive and then negative regulator lists |
<score 1>, <score 2>, …, <score \(k+l\)> |
examples |
Source list |
\(a^{\mathrm{sourcelist}}\) |
all sources of element’s positive and negative regulations from individual interactions, combined into a list, following the order of positive and then negative regulator lists |
<source 1>, <source 2>, …, <source \(k+l\)> |
examples |
Statements list |
\(a^{\mathrm{statementslist}}\) |
all support statements of element’s positive and negative regulations from individual interactions, combined into a list, following the order of positive and then negative regulator lists |
<statements 1>, <statements 2>, …, <statements \(k+l\)> |
examples |
Paper IDs list |
\(a^{\mathrm{paperIDslist}}\) |
all paper IDs (where statements are found) of element’s positive and negative regulations from individual interactions, combined into a list, following the order of positive and then negative regulator lists |
<paper IDs 1>, <paper IDs 2>, …, <paper IDs \(k+l\)> |
examples |
Whenever an individual regulator (positive or negative) has an empty attribute value, this is indicated with None in the list of attribute values.
Finally, several new model attributes are included in executable models to define element update rules, as well as element value and timing parameters for the simulation. These attributes are included under Simulation parameters in the figure above.
Attribute |
Symbol |
Description |
Values |
Examples |
|---|---|---|---|---|
Variable |
\(a^{\mathrm{variable}}\) |
Variable name used by simulators and other software |
<variable_name> |
examples |
Positive regulation rule |
\(a^{\mathrm{posregrule}}\) |
A rule used by simulation to compute the level of element’s positive regulation |
<string> //The rules for creating these positive regulation strings are written separately. |
examples |
Negative regulation rule |
\(a^{\mathrm{negregurule}}\) |
A rule used by simulation to compute the level of element’s negative regulation |
<string> //The rules for creating these negative regulation strings are written separately. |
examples |
Attribute |
Symbol |
Description |
Values |
Examples |
|---|---|---|---|---|
Value type |
\(a^{\mathrm{valuetype}}\) |
Type of value used for interpretting model outcomes |
|
examples |
Levels |
\(a^{\mathrm{levels}}\) |
Number of different levels (values) an element can be assigned; or infinite if a continuous variable |
<number of distinct levels> | |
examples |
State list number |
\(a^{\mathrm{statelist}}\) |
State list used in simulation to initialize the element and assign values throughout simulation; multiple state lists can be included and numbered, starting from 0 |
<value>,<value>[time],…,<value>[time] |
examples |
Const OFF |
\(a^{\mathrm{constOFF}}\) |
Indicates whether the element is assumed to be at the lowest allowed level (usually 0) for the entire simulation |
empty | \(\checkmark\) |
examples |
Const ON |
\(a^{\mathrm{constON}}\) |
Indicates whether the element is assumed to be at the highest allowed level for the entire simulation |
empty | \(\checkmark\) |
examples |
Increment |
\(a^{\mathrm{increment}}\) |
When the element can have more than two different levels, an increment indicates by how many levels it is allowed to increase or decrease in a single time step |
\(\Delta \mathrm{value}\) |
examples |
Attribute |
Symbol |
Description |
Values |
Examples |
|---|---|---|---|---|
Spontaneous |
\(a^{\mathrm{spontaneous}}\) |
Specifies spontaneous behavior of the element, if it has only positive or only negative regulators |
<non-negative integer> | |
examples |
Balancing |
\(a^{\mathrm{balancing}}\) |
Specifies the behavior of the element when its positive and negative regulation levels are equal |
{ |
examples |
Delay |
\(a^{\mathrm{delay}}\) |
description |
definition |
examples |
Update group |
\(a^{\mathrm{updategroup}}\) |
description |
definition |
examples |
Update rate |
\(a^{\mathrm{updaterate}}\) |
description |
definition |
examples |
Update rank |
\(a^{\mathrm{updaterank}}\) |
description |
definition |
examples |